Initiation of Replication
| Prokaryotic Replication | Table of Contents |
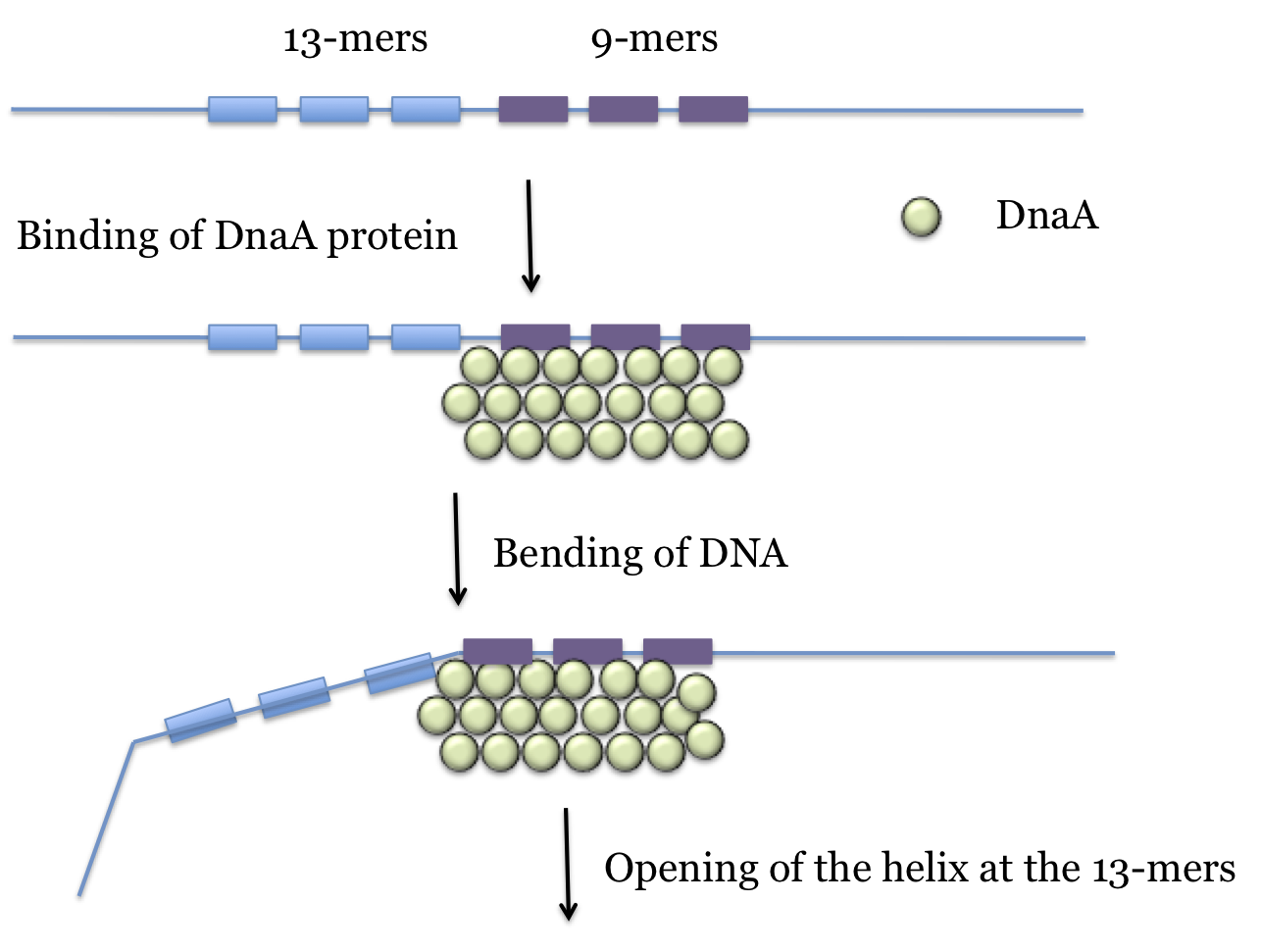
oriC:In E. coli replication initiates at a locus called oriC which is just a few hundred nucleotides in length. The main components are 3 copies of a 9-base sequence (9-mer) and 3 copies of a 13-base sequence (13-mer).
Formation of the pre-priming complex: About 20 copies of the DnaA protein bind to the 9-mer repeats of oriC. This complex results in a bending of the DNA that loosens the stability of the double helix. This results in the strands separating in the section of three 13-mers, forming what can be described as a "bubble" in the DNA.
At each end of the "bubble" a protein complex consisting of 6 copies each of DnaB and DnaC recognizes the opened complex, with the assistance of the DnaT protein. The 6 DnaB proteins of the complex bind to the DNA and the DnaC is released. The two strands of DNA fully separate at the site of the bend, a process that is assisted by DnaB which has helicase activity. This is the pre-priming complex. The DnaB that is bound will be part of the replication apparatus at the two growing forks that expand bidirectionally from the origin (one in each direction).
|
|
|
|
Priming: With the two strands now separated they can be used as templates. Since replication is bidirectional four primers need to be synthesized - one for each of the two strands in both directions. DNA primase binds to DnaB, primers are synthesized and DNA PolIII binds at each fork.
|
|
As the primers are extended we get into elongation.
| Prokaryotic Replication | Table of Contents |